
Exploring Pelagic Biodiversity of the Gulf of Alaska and the Impact of Its Seamounts
What Is DNA Barcoding And Why Is It Important?
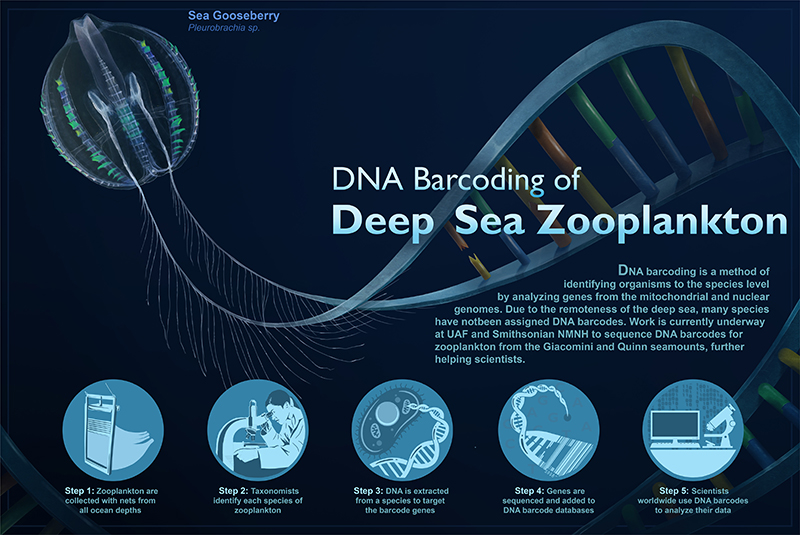
DNA barcoding is a way to identify a species by its genetic sequence. DNA from an organism is matched to a DNA barcode, which is a short fragment of DNA from a specific gene. A DNA barcode is like a fingerprint for a type of organism. DNA barcodes are used in a variety of ways, including to differentiate one species from another, identify larval or juvenile stages, and describe new species.
Download largest version (jpg, 2.88 MB).
A DNA barcode is like the barcode on something you’d buy at a store. When you scan an item, the checkout system searches a database for the matching barcode and tells you what the item is and other information, like how much it costs. With DNA, instead of a series of black and white bars, there is a series of letters (A, T, C, and G) that represent DNA bases — this is the genetic sequence. So, when a scientist checks a genetic sequence against a DNA barcode reference database, they’re able to identify the corresponding organism, assuming the organism is known and the barcode is in the database.
For more than 30 years, scientists have been generating DNA barcodes for all forms of life, from all around our planet to build DNA barcode reference databases. While substantial progress has been made, many of the organisms that call the ocean home have not been barcoded and may not even be known to science! This is due in part to the number, complexity, and diversity of species in our ocean — from tiny bacteria invisible to the naked eye to the blue whale, the largest animal on the planet. In addition, the deep ocean is the largest living space on Earth, yet it’s the least studied. So, there’s a lot of work remaining to be done.

Download largest version (jpg, 2.64 MB).
One of our objectives during the Exploring Pelagic Biodiversity of the Gulf of Alaska and the Impact of Its Seamounts expedition was to sample the pelagic environment of the deep ocean, focusing our efforts around two seamounts, Giacomini and Quinn. The expedition was heavily focused on the zooplankton community, which is an important link in marine food webs. During the expedition we collected zooplankton from the surface down to the bottom of the bathypelagic zone, with our deepest net tow reaching 4,000 meters (almost 2.5 miles).
Following net collections, our onboard team of zooplankton taxonomists worked diligently to pick through the samples, identify the many different species of zooplankton collected, and photograph, voucher, and catalog each organism, some of which were sent to the Smithsonian National Museum of Natural History’s Laboratories of Analytical Biology for DNA barcoding.

Download largest version (jpg, 3.59 MB).

Download largest version (jpg, 1.88 MB).
To date, over 1,000 organisms representing more than 200 species have been barcoded from the team’s 2019 expedition to the same seamounts. This expedition will add thousands more barcodes to the growing reference databases.
The need for DNA barcodes has become even more urgent given the promise of environmental sequencing: metabarcoding and environmental DNA (eDNA) and is emphasized in the recently released National Aquatic Environmental DNA Strategy. Environmental sequencing is a powerful tool for studying ecosystem biodiversity and monitoring environmental change and can be used to sample entire communities at different scales, by directly sampling animals collected in nets (metabarcoding) to collecting water samples containing genetic material shed by organisms into the water column (eDNA; e.g., mucus, feces, or tissue particles).
Analysis of environmental sequencing is a promising tool for monitoring biodiversity, but to be successful there must be something to match the genetic sequence against. This is where DNA barcode reference databases come into play. For the marine environment, MetaZooGene, SILVA, and PR2 are used most often to identify species from environmental samples using their DNA barcodes. However, these databases are far from complete, as many species have yet to be barcoded. As a result, many genetic datasets lack species-level identifications.
With so much work to be done, and so much to learn and discover, it’s an exciting time to be working collaboratively with taxonomists, molecular ecologists, and museum specialists to populate these incredibly important reference databases and shed more light on life in the deep ocean.
By Jennifer Questel, University of Alaska Fairbanks and Allen Collins, NOAA Fisheries’ National Systematics Laboratory and Smithsonian National Museum of Natural History
