
The octocorals Metallogorgia sp. (center) and Paramuricea sp. (lower left) on Kelvin Seamount. Click image for larger view.
Seamount Coral Genetics
Scott C. France, PhD
Assistant Professor
Department of Biology
University of Louisiana at Lafayette
Co-Principal Investigator
Mountains in the Sea 2004
It is no accident that the Galapagos Islands are closely associated with Charles Darwin's theory of evolution. The organisms living on these oceanic islands are hundreds of miles away from the nearest mainland, isolated by a deep expanse of ocean. When populations of organisms become geographically isolated, they cannot reproduce with other individuals of the species. Gradually, they evolve in separate ways. This explains why isolated island chains (such as Galapagos and Hawaii) and large islands (such as New Zealand) have so many species not found elsewhere. These geographically-isolated species are called "endemics."
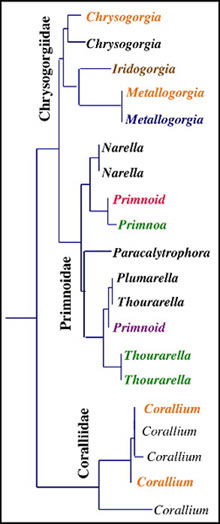
This phylogenetic tree shows the relationships among coral species from three families (Chrysogorgiidae, Primnoidae, Coralliidae) based on DNA sequences of the msh1 gene. The longer the branch separating corals, the greater the genetic divergence between them. Click image for larger view and more details.
Seamounts are the deep-sea equivalent of oceanic islands. Rising thousands of meters above the surrounding sea floor, these peaks create islands of hard substrate in the water column. Our question is: are the species inhabiting deep seamounts endemic, or are they simply populations of species with broader geographic distributions? We are addressing this question through the use of DNA-sequencing technology.
Every cell of living organisms contains DNA, the molecule that holds the instructions for building the organism. A DNA molecule has two intertwined strands, each essentially a string of smaller molecules, called "nucleotides." The specific order of the nucleotides along the strand contains information, just as the order of letters in a word has a specific meaning. Rearrange the letters in the word “lamp,” and you can get a word with an entirely new meaning: “palm.” Genes are the same: they are chunks of DNA that code for specific products. One change in the order of the nucleotides in a gene may code for a protein that produces hair, while another produces scales. Today, thanks to the rapid progress in biotechnology over the last 20 years, we can read the sequence of nucleotides that comprise DNA strands. Since every species differs somewhat from all others, their DNA sequences also differ. Scientists have suggested DNA sequences can be used as a "barcode" to identify species.
As part of Mountains in the Sea 2004, we are collecting tissue samples from seamount corals, sequencing portions of their DNA, and comparing the DNA sequences to determine the evolutionary relationships among the corals. Our analyses have two major objectives:
1.)We aim to reveal the existence of morphologically cryptic species, which are species that look so similar that it is nearly impossible to distinguish between them. It is fairly easy to identify most species based on their morphology. For example, the octocoral Metallogorgia looks like a tall, pink palm tree, whereas Paramuricea sp. is a small, orange fan. But species that have diverged relatively recently, in evolutionary terms, may reveal little or no morphological differentiation that shows they are reproductively isolated. Changes in DNA, however, accumulate more rapidly than do morphological changes. By comparing DNA sequences, we may discover coral species previously unknown to science that are morphologically similar but genetically very different. This will give us a better understanding species diversity (the number of species) on seamounts and their potential as centers of speciation.

After DNA is isolated from the coral tissue, a series of biochemical reactions leads to this output from a DNA Sequencer. Here the sequence of nucleotides that encode the gene can be read from left to right. Click image for larger view.
2.) We also plan to determine whether populations of a single species, distributed across multiple seamounts, are isolated. Even individuals of the same species are genetically variable. The more closely individuals are related, the fewer differences will be seen when comparing their DNA sequences. For instance, siblings will show very little divergence in their DNA, whereas distant relatives will show more. The longer the time since two individuals shared a common ancestor, the more differences we should observe when comparing their DNA. (This same principle underlies the use of DNA for identification in criminal forensics cases.)
We can use this principle to predict how long populations of a species have been isolated from one another. For example, consider the Metallogorgia we observed on multiple seamounts during our Mountains in the Sea 2003 cruise. Are the populations living on Kelvin Seamount completely isolated from those living on Manning Seamount, or do their larvae "routinely" travel across the deep water that separates them? High levels of genetic divergence between populations on the different seamounts would suggest that little exchange occurs between seamounts. If we see seamount populations are completely genetically isolated, we would predict that larvae will not colonize from one seamount to another. This information may be useful from a practical standpoint, with important conservation implications.
If deep-sea coral larvae can travel between seamounts, can they also disperse from seamounts to the North American continental shelf? We have been sampling the corals of the deep Gulf of Maine and canyons of Georges Bank (Deep East 2001) and are comparing their DNA sequences to evaluate evolutionary relationships.
During Mountains of the Sea 2003, we sampled 20 coral species for genetics, including six black corals and one stony coral. Our observations indicated that some species (e.g., Iridogorgia sp., Metallogorgia sp., and Paramuricea sp.) were more widespread and abundant across the seamount chain. On this year’s expedition, we will collect tissue fragments from more individuals of these species to get an appropriate sample size for population genetic analysis. Will we find several different species of each genus, or patterns suggesting high levels of larval dispersal between seamounts? These studies will be directly linked to those of Larval Settlement. If we observe new recruits, their DNA may tell us where they came from.





















