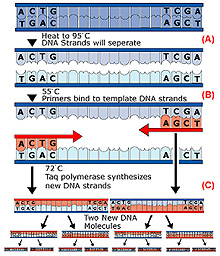
An illustration of the polymerase chain reaction (PCR). Step 1: Solution is heated to 95°C to denature ("unzip") the two strands of the target DNA (A). Step 2: Solution is cooled to ~55°C to allow the primers to anneal (bind) to the ends of the DNA strands (B). Step 3: Solution is reheated to ~75°C to allow TAQ polymerase to synthesize complementary copies of each strand. Click image for larger view.
How Do Scientists Analyze Coral DNA?
Marion
Beal
Geneticist
South Carolina Department of Natural Resources
Genetics Laboratory
Hollings Marine Lab
The analysis of DNA isolated from deep-water corals can provide information on the relationships among coral colonies of the same species as well as the evolutionary relationship among different species of corals. Genetic analysis can also tell us how corals reproduce: sexual reproduction vs. colony fragmentation and growth of fragments. This analysis can help us understand how corals are dispersed to new habitats (e.g., seamounts) as they are formed, or how corals might recruit to an area where they have previously been eliminated by natural or man-made destruction.
Scientists use a variety of techniques to analyze DNA.
Population genetics
We know very little about the life history of deep-sea coral species. Fortunately, population genetics offers us a useful tool for gaining knowledge on reproduction
and dispersal. The relatively sedentary life of adult coral, and
reproduction of colonies by fragmentation, can lead to populations or
groups of colonies that are genetically isolated from other populations. However,
sexual reproduction by broadcast spawning and dispersal of tiny free-swimming
larvae provide mechanisms of gene flow for corals that may result in
geographically distant populations being similar in genetic structure.
Variability in the sequence of base pairs in the DNA molecule allows us to identify genetic differences that may accumulate between individuals and populations, and to define the historical or evolutionary relationships based on these traits. Genetic data may also be used to help understand biological, oceanographic, and geographic factors that isolate populations and lead to genetic and morphological differentiation.
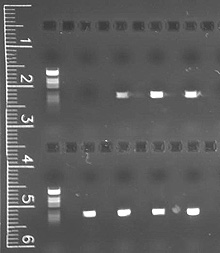
This image shows how we "visualize" DNA through the agarose gel electrophoresis of polymerase chain reaction product. Click image for larger view.
DNA collection
Collection and storage of DNA has become relatively simple. New technology
allows us to obtain analyzable DNA from only a small
tissue sample that can be preserved using simple methods. We can place small
tissue samples in preservative solutions
and store them at ambient temperature, and the DNA in the tissue will remain
stable for several years. After
isolation from the tissue, the DNA can be further stabilized
and stored dry.
The first step in DNA analysis is to isolate the DNA from the other organic materials in the tissue sample. Isolation methods take advantage of the differences in molecular size and structure between DNA and proteins, lipids, and other large molecules in the cells from which the DNA is extracted.
Most analysis techniques require very small amounts of DNA, and one sample can be used for several different analyses. Microsatellite and mitochondrial DNA (mtDNA) are commonly used for analysis, because they mutate rapidly and are highly variable regions of the DNA. In addition, variation in the sequence of bases in the DNA and the resulting allelic variations are easily revealed with fairly simple molecular techniques.
Using replication to "stretch" a sample
DNA can replicate itself in a cell, and geneticists use this characteristic
to produce many copies of the DNA in laboratory reactions from a small
tissue sample. The polymerase chain reaction (PCR) is used to produce
millions of copies of a specific section of DNA that has been isolated
from the tissue sample. Amplification takes place in a matter of
hours.
During that time, a reaction containing a pair of short-priming DNA sequences, complementary to the ends of the target sequence, TAQ DNA polymerase, and a solution of deoxynucleotides (dNTPs), is repeatedly heated and cooled. This denatures the double-stranded DNA, synthesizes a new strand (to complement each of the denatured strands), and repeats the cycle.
In order to visualize the DNA, we load the amplified PCR product into a gel matrix. When we apply an electrical current to the gel, the DNA migrates through the gel at a rate that depends on fragment size. After the run, DNA trapped in the gel can be visualized using the chemical ethidium bromide, which integrates into the DNA and fluoresces under ultraviolet light.

A researcher examines an image of a gel for the presence of microsatellite DNA, indicated by the repeating pattern of bands. Click image for larger view.
Digesting the DNA
After confirming the presence of amplified DNA (of a size that agrees
with the size of the sequence targeted for amplification), the amplified
mtDNA can be digested with restriction enzymes. Restriction enzymes
recognize certain sequences of DNA and cut the DNA into pieces at those
sequences. A population that possesses a mutation in the DNA(so
that it does not have that particular sequence) will end up post-digestion with DNA
fragments that are different from other populations.
When the amplified DNA that has been subject to digestion reactions is subjected to gel electrophoresis, smaller fragments of DNA travel farther in the gel than larger fragments, thus revealing variations in the sequence of base pairs in the amplified DNA. It also reveals differences that may reflect species or population differences among the samples of coral from which the DNA was obtained.
Microsatellite DNA amplified by PCR is not subjected to restriction digestion, but may differ among populations in the size of the microsatellite. Again, because of different sizes of the DNA molecule in different-sized microsatellites, the microsatellite's DNA will migrate through the gel at different rates, based on size of the sequence amplified. Microsatellite size varies among populations, and the resulting gel can reveal population differences based on the frequency of microsatellites of differing sizes.
An even closer examination
For an even closer examination of variations, variable regions within
the mtDNA or microsatellite DNA can be sequenced from PCR products, revealing
single nucleotide differences between individual sequences.

This researcher is loading a DNA sample into a gel. The gel will be subjected to an electric current, which will cause the DNA molecules to move through the gel. The distance and speed they move is related to the size of the DNA fragments, which varies from species to species, or among populations. Click image for larger view.
The data on the frequency of detected sequence differences among individual samples (coral colonies) can then be statistically tested with software designed for molecular genetic analysis.
Large numbers of alleles are indicative of high genetic variability in a population; and large numbers of shared alleles among populations is evidence for gamete or larval exchange between communities. On the other hand, small numbers of alleles indicate low genetic variability or possible “inbreeding”; and a low number of shared alleles between populations may be due to poor dispersal of larvae or gametes.
Saving samples for later testing
The DNA samples, whether from corals or other organisms, can be archived and stored
for future research. The samples can be easily exchanged among investigators. Molecular technologies are advancing
rapidly and leading to new ways to analyze DNA. In the near future,
there could be endless possibilities for analysis of
samples collected today.